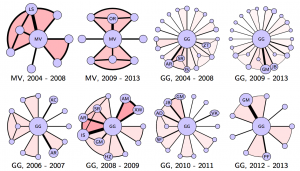
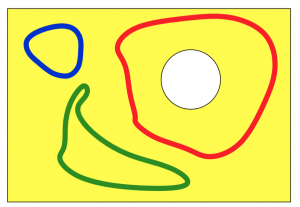
Data is getting big, but more than big it is getting pervasive. As our lives become more dependent and integrated with the digital infrastructure, more aspects of our life get measured and recorded. This pervasiveness leads to the emergence of novel types of signals for which existing analytic tools cannot be applied easily. Networked data falls in this category. As an example, the networks above are the co-authorship networks constructed from IEEE Transaction on Signal Processing publications of two famous signal processing researchers, Prof. Georgios B. Giannakis (GG) of the University of Minnesota and Prof. Martin Vetterli (MV) of the École Polytechnique Fédérale de Lausannee for the selected timeframe. Each node in the networks represents an author. The size of the nodes is proportional to the number of publications of the corresponding authors. The width of the links is proportional to the number of publications between the author pairs. Color intensity of the shading triangles is proportional to the number of publications to the enclosed author triplets. Distinguishable differences exist in their respective collaboration patterns. A natural question is: how can we quantify the differences between high order networks?
Such a problem has been solved by comparing some features of the respective networks. Feature analysis is application dependent, utilizes only a small portion of the information conveyed, and may yield conflicting judgments — two networks close to each other are quiet different from the third. For the networks presented above, it is not clear which features one should use. Moreover, feature analysis is not easily generalizable to incorporate high dimensional information, e.g. publications between three or four authors, as what we did in the example. We tackle this problem from a different perspective — by defining valid metric distances, which are universal, depend on all edge weights, and are null if and only if the networks are isomorphic. We also establish a connection between Network Science and Computational Topology and uses tools from the latter to efficiently quantify network differences.
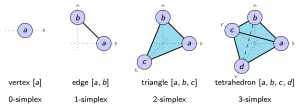
In specific, besides relationship between pairs of nodes, we also consider high order relationships between three nodes or four nodes. Such consideration is meaningful in practice, because the collaboration between three authors is different from the collaboration between two authors, and collaboration between four authors is not the same as the collaboration between two authors. Formally, we consider high order networks of order $K$ as a tuple $N_X^K = (X, r_X^0, r_X^1, \dots, r_X^K)$, where $X$ is the set of nodes, and $r_X^k$ is a mapping from $k+1$ tuples to $\mathbb{R}_+$ quantifying the relationship between $k+1$ elements in $X$. We require that the relationship $r_X^2(A, B, B)$ between $A$, $B$, and $B$ to be the same as the relationship $r_X^1(A, B)$ between $A$ and $B$. In common scenarios, relationship either quantifies the similarity or the dissimilarity between tuples. When it denotes proximities, we require $r_X^2(A, B, C) \leq r_X^1(A, B)$; when relationship functions represent dissimilarity, we require $r_X^2(A, B, C) \geq r_X^1(A, B)$.
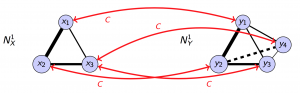
Because we need to measure the dissimilarity between networks of arbitrary sizes, we need a way to map nodes between networks with different number of nodes. We use a mathematical tool called correspondence. A correspondence $C$ between two set of nodes $X$ and $Y$ is a subset of $X \times Y$ such that for any $x \in X$, there exists $y \in Y$ with $(x, y) \in C$, and for any $y \in Y$, there exists $x \in X$ with $(x, y) \in C$. This is a generalization of permutation and enables us to map node sets with different cardinality. Given a pair of networks $N_X^K$ and $N_Y^K$, and a specific correspondence $C$, the difference between the two networks measured by the correspondence can be defined as the bottleneck tuple where the respective relationship functions is the most different
\begin{align}
\Gamma_{X, Y}^k (C) = \max_{(x_{0:k}, y_{0:k}) \in C} \left\| r_X^k(x_{0:k} – r_Y^k(y_{0:k}))\right\|.
\end{align}
The network distance between the pair of networks for the $k$-th order can then be defined by searching for the correspondence that makes the networks the most similar,
\begin{align}
d_{\mathcal{N}}^k (N_X^K, N_Y^K) = \min_{C \in \mathcal{C}(X, Y)} \left\{ \Gamma_{X, Y}^k (C) \right\}.
\end{align}
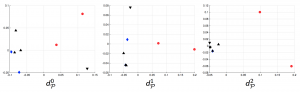

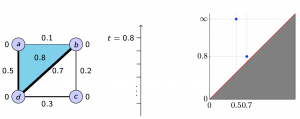
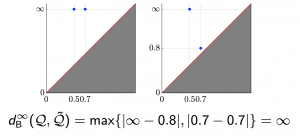
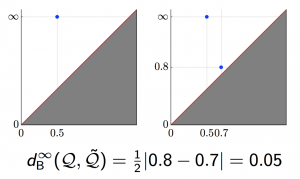
We applied these homological lower bounds to distinguish networks constructed from publications in different journals, e.g. engineering and mathematics. We found that the network lower bounds could successfully distinguish the research patterns in different communities. We also considered a more challenging task, to distinguish networks from engineering research communities with different interest, e.g. signal processing, automatic control, and wireless communication. These homological tools made a moderate success in classifying them as well.
